Plotting in Makie
Plotting in Makie works somewhat differently than Plots, since the recipe system is different. You can pass a 2-D raster to any surface-like function (heatmap, contour, contourf, or even surface for a 3D plot) with ease.
2-D rasters in Makie
using CairoMakie, Makie
using Rasters, RasterDataSources, ArchGDAL
A = Raster(WorldClim{BioClim}, 5) # this is a 3D raster, so is not accepted.┌ 2160×1080 Raster{Union{Missing, Float32}, 2} bio5 ┐
├───────────────────────────────────────────────────┴──────────────────── dims ┐
↓ X Projected{Float64} -180.0:0.16666666666666666:179.83333333333331 ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} 89.83333333333333:-0.16666666666666666:-90.0 ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 1 entry:
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
├────────────────────────────────────────────────────────────────────── raster ┤
missingval: missing
extent: Extent(X = (-180.0, 179.99999999999997), Y = (-90.0, 90.0))
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.25722...
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 … -89.6667 -89.8333 -90.0
-180.0 missing missing missing -15.399 -13.805 -14.046
-179.833 missing missing missing -15.9605 -14.607 -14.5545
⋮ ⋱ ⋮
179.667 missing missing missing -18.2847 -16.7513 -16.72
179.833 missing missing missing … -17.1478 -15.4243 -15.701fig, ax, _ = plot(A)
contour(fig[1, 2], A)
ax = Axis(fig[2, 1]; aspect = DataAspect())
contourf!(ax, A)
surface(fig[2, 2], A) # even a 3D plot work!
fig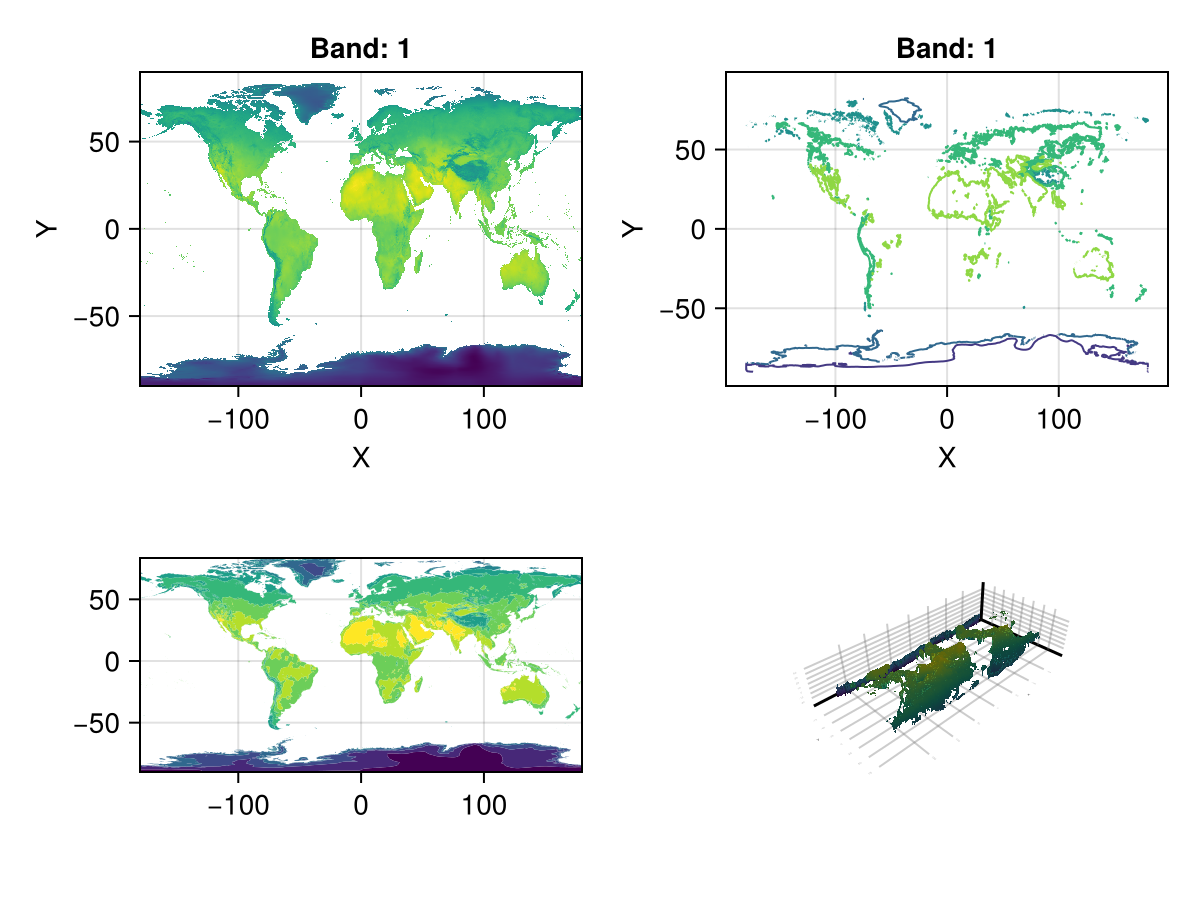
3-D rasters in Makie
Warning
This interface is experimental, and unexported for that reason. It may break at any time!
Just as in Plots, 3D rasters are treated as a series of 2D rasters, which are tiled and plotted.
You can use Rasters.rplot to visualize 3D rasters or RasterStacks in this way. An example is below:
stack = RasterStack(WorldClim{Climate}; month = 1)
Rasters.rplot(stack; Axis = (aspect = DataAspect(),),)
You can pass any theming keywords in, which are interpreted by Makie appropriately.
Plotting with Observables, animations
Rasters.rplot should support Observable input out of the box, but the dimensions of that input must remain the same - i.e., the element names of a RasterStack must remain the same.
# `stack` is the WorldClim climate data for January
stack_obs = Observable(stack)
fig = Rasters.rplot(stack_obs;
Colorbar=(; height=Relative(0.75), width=5)
)
record(fig, "rplot.mp4", 1:12; framerate = 3) do i
stack_obs[] = RasterStack(WorldClim{Climate}; month = i)
end"rplot.mp4"Makie.set_theme!() # reset themeRasters.rplot Function
Rasters.rplot([position::GridPosition], raster; kw...)raster may be a Raster (of 2 or 3 dimensions) or a RasterStack whose underlying rasters are 2 dimensional, or 3-dimensional with a singleton (length-1) third dimension.
Keywords
plottype = Makie.Heatmap: The type of plot. Can be any Makie plot type which accepts aRaster; in practice,Heatmap,Contour,ContourfandSurfaceare the best bets.axistype = Makie.Axis: The type of axis. This can be anAxis,Axis3,LScene, or even aGeoAxisfrom GeoMakie.jl.X = XDim: The X dimension of the raster.Y = YDim: The Y dimension of the raster.Z = YDim: The Y dimension of the raster.draw_colorbar = true: Whether to draw a colorbar for the axis or not.colorbar_position = Makie.Right(): Indicates which side of the axis the colorbar should be placed on. Can beMakie.Top(),Makie.Bottom(),Makie.Left(), orMakie.Right().colorbar_padding = Makie.automatic: The amount of padding between the colorbar and its axis. Ifautomatic, then this is set to the width of the colorbar.title = Makie.automatic: The titles of each plot. Ifautomatic, these are set to the name of the band.xlabel = Makie.automatic: The x-label for the axis. Ifautomatic, set to the dimension name of the X-dimension of the raster.ylabel = Makie.automatic: The y-label for the axis. Ifautomatic, set to the dimension name of the Y-dimension of the raster.colorbarlabel = "": Usually nothing, but here if you need it. Sets the label on the colorbar.colormap = nothing: The colormap for the heatmap. This can be set to a vector of colormaps (symbols, strings,cgrads) if plotting a 3D raster or RasterStack.colorrange = Makie.automatic: The colormap for the heatmap. This can be set to a vector of(low, high)if plotting a 3D raster or RasterStack.nan_color = :transparent: The color whichNaNvalues should take. Default to transparent.
Using vanilla Makie
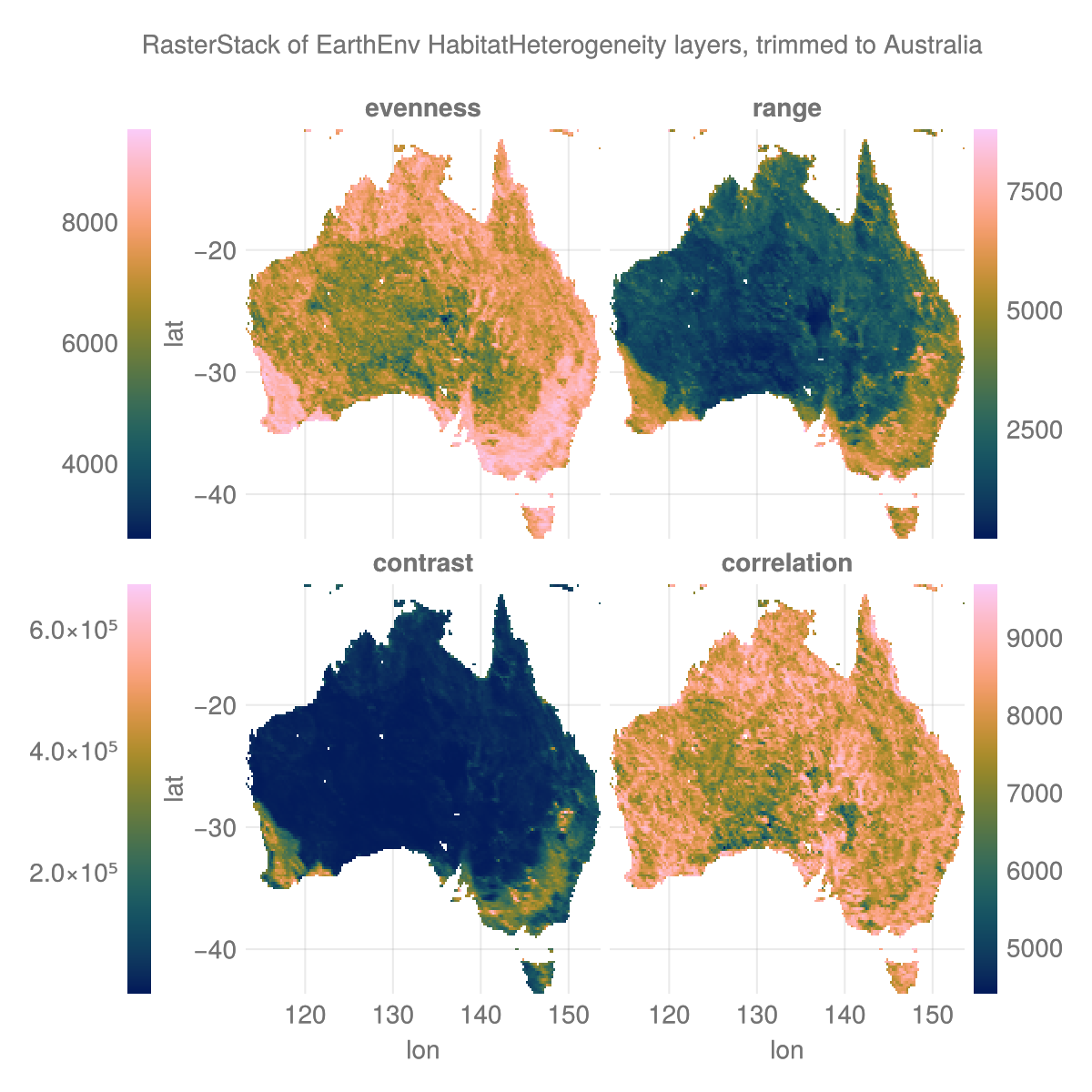
using Rasters, RasterDataSourcesThe data
layers = (:evenness, :range, :contrast, :correlation)
st = RasterStack(EarthEnv{HabitatHeterogeneity}, layers)
ausbounds = X(100 .. 160), Y(-50 .. -10) # Roughly cut out australia
aus = st[ausbounds...] |> Rasters.trim┌ 194×161 RasterStack ┐
├─────────────────────┴────────────────────────────────────────────────── dims ┐
↓ X Projected{Float64} 113.33333333333334:0.20833333333333334:153.54166666666669 ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} -10.208333333333334:-0.20833333333333334:-43.541666666666664 ReverseOrdered Regular Intervals{Start}
├────────────────────────────────────────────────────────────────────── layers ┤
:evenness eltype: Union{Missing, UInt16} dims: X, Y size: 194×161
:range eltype: Union{Missing, UInt16} dims: X, Y size: 194×161
:contrast eltype: Union{Missing, UInt32} dims: X, Y size: 194×161
:correlation eltype: Union{Missing, UInt16} dims: X, Y size: 194×161
├────────────────────────────────────────────────────────────────────── raster ┤
missingval: missing
extent: Extent(X = (113.33333333333334, 153.75000000000003), Y = (-43.541666666666664, -10.0))
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.25722...
└──────────────────────────────────────────────────────────────────────────────┘The plot
# colorbar attributes
colormap = :batlow
flipaxis = false
tickalign=1
width = 13
ticksize = 13
# figure
with_theme(theme_dark()) do
fig = Figure(; size=(600, 600), backgroundcolor=:transparent)
axs = [Axis(fig[i,j], xlabel = "lon", ylabel = "lat",
backgroundcolor=:transparent) for i in 1:2 for j in 1:2]
plt = [Makie.heatmap!(axs[i], aus[l]; colormap) for (i, l) in enumerate(layers)]
for (i, l) in enumerate(layers) axs[i].title = string(l) end
hidexdecorations!.(axs[1:2]; grid=false, ticks=false)
hideydecorations!.(axs[[2,4]]; grid=false, ticks=false)
Colorbar(fig[1, 0], plt[1]; flipaxis, tickalign, width, ticksize)
Colorbar(fig[1, 3], plt[2]; tickalign, width, ticksize)
Colorbar(fig[2, 0], plt[3]; flipaxis, tickalign, width, ticksize)
Colorbar(fig[2, 3], plt[4]; tickalign, width, ticksize)
colgap!(fig.layout, 5)
rowgap!(fig.layout, 5)
Label(fig[0, :], "RasterStack of EarthEnv HabitatHeterogeneity layers, trimmed to Australia")
fig
end
save("aus_trim.png", current_figure());