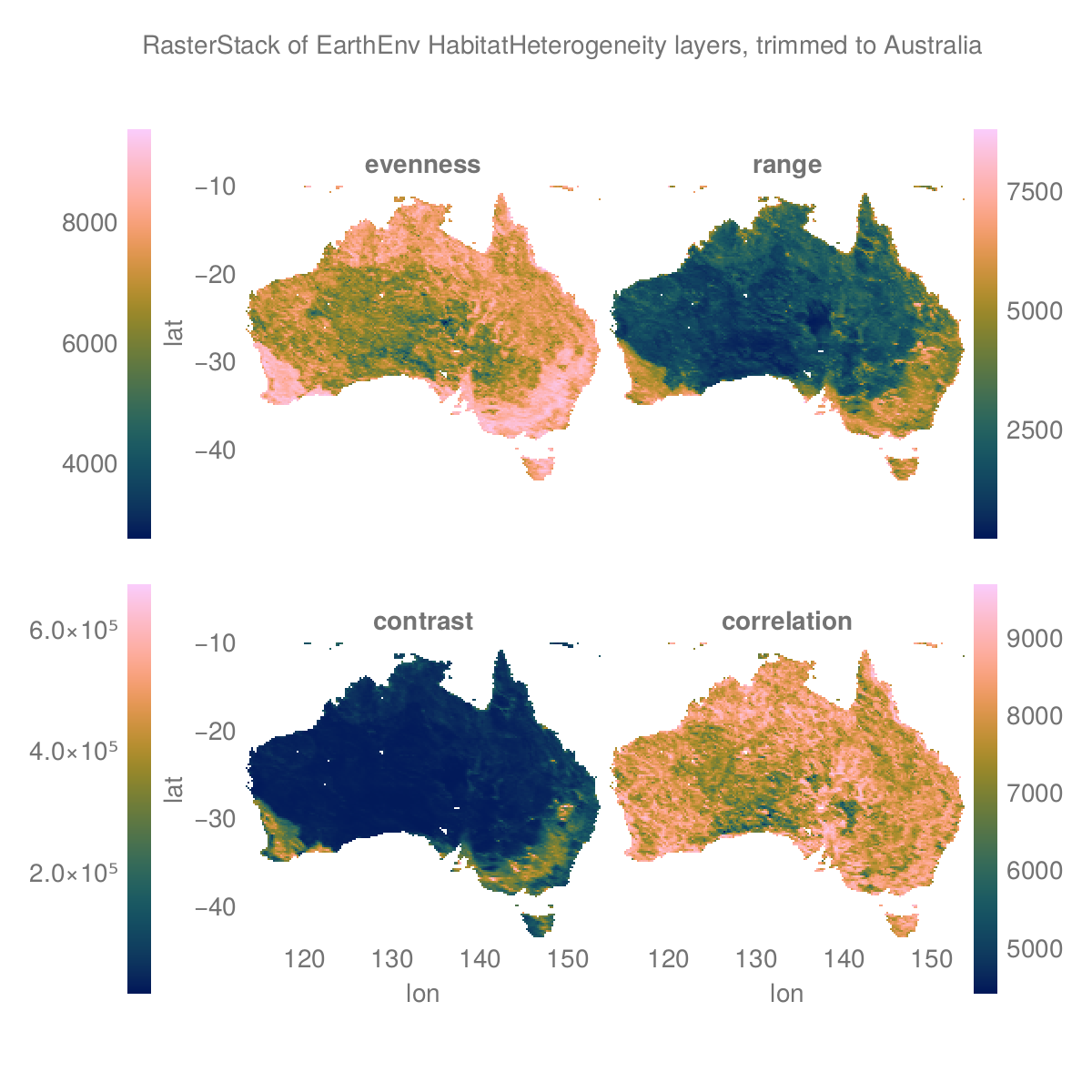
Most base methods work as for regular julia Arrays, such as reverse and rotations like rotl90. Base, statistics and linear algebra methods like mean that take a dims argument can also use the dimension name.
Mean over the time dimension:
using Rasters, Statistics, RasterDataSources
A = Raster(WorldClim{BioClim}, 5)╭──────────────────────────────────╮
│ 2160×1080 Raster{Float32,2} bio5 │
├──────────────────────────────────┴───────────────────────────────────── dims ┐
↓ X Projected{Float64} LinRange{Float64}(-180.0, 179.83333333333331, 2160) ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(X = (-180.0, 179.99999999999997), Y = (-90.0, 90.0))
missingval: -3.4f38
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 89.3333 … -89.6667 -89.8333 -90.0
-180.0 -3.4f38 -3.4f38 -3.4f38 -3.4f38 -15.399 -13.805 -14.046
⋮ ⋱ ⋮
179.833 -3.4f38 -3.4f38 -3.4f38 -3.4f38 … -17.1478 -15.4243 -15.701Then we do the mean over the X dimension
mean(A, dims=X) # Ti if time were available would also be possible╭───────────────────────────────╮
│ 1×1080 Raster{Float32,2} bio5 │
├───────────────────────────────┴──────────────────────────────────────── dims ┐
↓ X Projected{Float64} -180.0:360.0:-180.0 ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(X = (-180.0, 180.0), Y = (-90.0, 90.0))
missingval: -3.4f38
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 89.3333 … -89.6667 -89.8333 -90.0
-180.0 -Inf -Inf -Inf -Inf -23.0768 -22.9373 -22.0094broadcast works lazily from disk when lazy=true, and is only applied when data is directly indexed. Adding a dot to any function will use broadcast over a Raster just like an Array.
Broadcasting
For a disk-based array A, this will only be applied when indexing occurs or when we read the array.
A .*= 2╭──────────────────────────────────╮
│ 2160×1080 Raster{Float32,2} bio5 │
├──────────────────────────────────┴───────────────────────────────────── dims ┐
↓ X Projected{Float64} LinRange{Float64}(-180.0, 179.83333333333331, 2160) ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(X = (-180.0, 179.99999999999997), Y = (-90.0, 90.0))
missingval: -3.4f38
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 … -89.6667 -89.8333 -90.0
-180.0 -Inf -Inf -Inf -30.798 -27.61 -28.092
⋮ ⋱ ⋮
179.833 -Inf -Inf -Inf … -34.2955 -30.8485 -31.402To broadcast directly to disk, we need to open the file in write mode:
open(Raster(filename); write=true) do O
O .*= 2
endTo broadcast over a RasterStack use map, which applies a function to the raster layers of the stack.
newstack = map(stack) do raster
raster .* 2
endModifying object properties
rebuild can be used to modify the fields of an object, generating a new object (but possibly holding the same arrays or files).
If you know that a file had an incorrectly specified missing value, you can do:
rebuild
A = Raster(WorldClim{BioClim}, 5)
rebuild(A; missingval=-9999)╭──────────────────────────────────╮
│ 2160×1080 Raster{Float32,2} bio5 │
├──────────────────────────────────┴───────────────────────────────────── dims ┐
↓ X Projected{Float64} LinRange{Float64}(-180.0, 179.83333333333331, 2160) ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(X = (-180.0, 179.99999999999997), Y = (-90.0, 90.0))
missingval: -9999.0f0
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 89.3333 … -89.6667 -89.8333 -90.0
-180.0 -3.4f38 -3.4f38 -3.4f38 -3.4f38 -15.399 -13.805 -14.046
⋮ ⋱ ⋮
179.833 -3.4f38 -3.4f38 -3.4f38 -3.4f38 … -17.1478 -15.4243 -15.701(replace_missing will actually replace the current values).
Or if you need to change the name of the layer:
Then use rebuild as
B = rebuild(A; name=:temperature)
B.name:temperaturereplace_missing
replace_missing(A, missingval=-9999)╭──────────────────────────────────╮
│ 2160×1080 Raster{Float32,2} bio5 │
├──────────────────────────────────┴───────────────────────────────────── dims ┐
↓ X Projected{Float64} LinRange{Float64}(-180.0, 179.83333333333331, 2160) ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(X = (-180.0, 179.99999999999997), Y = (-90.0, 90.0))
missingval: -9999.0f0
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 … -89.6667 -89.8333 -90.0
-180.0 -9999.0 -9999.0 -9999.0 -15.399 -13.805 -14.046
⋮ ⋱ ⋮
179.833 -9999.0 -9999.0 -9999.0 … -17.1478 -15.4243 -15.701set
set can be used to modify the nested properties of an objects dimensions, that are more difficult to change with rebuild. set works on the principle that dimension properties can only be in one specific field, so we generally don't have to specify which one it is. set will also try to update anything affected by a change you make.
This will set the X axis to specify points, instead of intervals:
using Rasters: Points
set(A, X => Points)╭──────────────────────────────────╮
│ 2160×1080 Raster{Float32,2} bio5 │
├──────────────────────────────────┴───────────────────────────────────── dims ┐
↓ X Projected{Float64} LinRange{Float64}(-180.0, 179.83333333333331, 2160) ForwardOrdered Regular Points,
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(X = (-180.0, 179.83333333333331), Y = (-90.0, 90.0))
missingval: -3.4f38
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 89.3333 … -89.6667 -89.8333 -90.0
-180.0 -3.4f38 -3.4f38 -3.4f38 -3.4f38 -15.399 -13.805 -14.046
⋮ ⋱ ⋮
179.833 -3.4f38 -3.4f38 -3.4f38 -3.4f38 … -17.1478 -15.4243 -15.701We can also reassign dimensions, here X becomes Z:
set(A, X => Z)╭──────────────────────────────────╮
│ 2160×1080 Raster{Float32,2} bio5 │
├──────────────────────────────────┴───────────────────────────────────── dims ┐
↓ Z Projected{Float64} LinRange{Float64}(-180.0, 179.83333333333331, 2160) ForwardOrdered Regular Intervals{Start},
→ Y Projected{Float64} LinRange{Float64}(89.83333333333333, -90.0, 1080) ReverseOrdered Regular Intervals{Start}
├──────────────────────────────────────────────────────────────────── metadata ┤
Metadata{Rasters.GDALsource} of Dict{String, Any} with 4 entries:
"units" => ""
"offset" => 0.0
"filepath" => "./WorldClim/BioClim/wc2.1_10m_bio_5.tif"
"scale" => 1.0
├────────────────────────────────────────────────────────────────────── raster ┤
extent: Extent(Z = (-180.0, 179.99999999999997), Y = (-90.0, 90.0))
missingval: -3.4f38
crs: GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
└──────────────────────────────────────────────────────────────────────────────┘
↓ → 89.8333 89.6667 89.5 89.3333 … -89.6667 -89.8333 -90.0
-180.0 -3.4f38 -3.4f38 -3.4f38 -3.4f38 -15.399 -13.805 -14.046
⋮ ⋱ ⋮
179.833 -3.4f38 -3.4f38 -3.4f38 -3.4f38 … -17.1478 -15.4243 -15.701setcrs(A, crs) and setmappedcrs(A, crs) will set the crs value/s of an object to any GeoFormat from GeoFormatTypes.jl.